User Interaction With The Plot Window¶
The class pymassspec_plot.ClickEventHandler allows for additional interaction with
the plot on top of that provided by matplotlib.
Note
This may not work in Jupyter Notebook.
To use the class, first import and process the data as before:
from domdf_python_tools.paths import PathPlus
import matplotlib.pyplot as plt
from pyms.GCMS.IO.JCAMP import JCAMP_reader
from pyms.IntensityMatrix import build_intensity_matrix
from pyms.Display import plot_ic, plot_peaks
from pyms.Noise.SavitzkyGolay import savitzky_golay
from pyms.TopHat import tophat
from pyms.BillerBiemann import BillerBiemann, rel_threshold, num_ions_threshold
cwd = PathPlus(".").resolve()
data_directory = PathPlus(".").resolve().parent.parent / "datafiles"
# Change this if the data files are stored in a different location
output_directory = cwd / "output"
jcamp_file = data_directory / "gc01_0812_066.jdx"
data = JCAMP_reader(jcamp_file)
data.trim("500s", "2000s")
tic = data.tic
im = build_intensity_matrix(data)
-> Reading JCAMP file '/home/runner/work/PyMassSpec-Plot/PyMassSpec-Plot/datafiles/gc01_0812_066.jdx'
Trimming data to between 520 and 4517 scans
n_scan, n_mz = im.size
for ii in range(n_mz):
ic = im.get_ic_at_index(ii)
ic_smooth = savitzky_golay(ic)
ic_bc = tophat(ic_smooth, struct="1.5m")
im.set_ic_at_index(ii, ic_bc)
peak_list = BillerBiemann(im, points=9, scans=2)
pl = rel_threshold(peak_list, percent=2)
new_peak_list = num_ions_threshold(pl, n=3, cutoff=10000)
print("Number of filtered peaks: ", len(new_peak_list))
Number of filtered peaks: 72
Creating the plot proceeds much as before, except that
pymassspec_plot.ClickEventHandler must be called before
show().
You should also assign this to a variable to prevent it being garbage collected.
from pyms.Display import ClickEventHandler
fig, ax = plt.subplots(1, 1, figsize=(8, 5))
# Plot the TIC
plot_ic(ax, tic, label="TIC")
# Plot the peaks
plot_peaks(ax, new_peak_list)
# Set the title
ax.set_title('TIC for gc01_0812_066 with Detected Peaks')
# Set up the ClickEventHandler
handler = ClickEventHandler(new_peak_list)
# Add the legend
plt.legend()
plt.show()
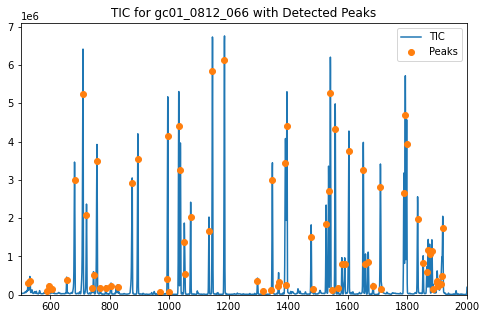
Clicking on a peak causes a list of the 5 highest intensity ions at that peak to be written to the terminal in order.
The output should look similar to this:
RT: 1031.823
Mass Intensity
158.0 2206317.857142857
73.0 628007.1428571426
218.0 492717.04761904746
159.0 316150.4285714285
147.0 196663.95238095228
If there is no peak close to the point on the chart that was clicked, the following will be shown in the terminal:
No Peak at this point
The pymassspec_plot.ClickEventHandler class can be configured with a different
tolerance, in seconds, when clicking on a Peak, and to display a
different number of top n ions when a Peak is clicked.
In addition, clicking the right mouse button on a Peak displays the mass spectrum at the peak in a new window.

Fig. 5 The mass spectrum displayed by PyMassSpec-Plot when a peak in the graphics window is right clicked¶
To zoom in on a portion of the plot, select the  button,
hold down the left mouse button while dragging a rectangle over the area of interest.
To return to the original view, click on the
button,
hold down the left mouse button while dragging a rectangle over the area of interest.
To return to the original view, click on the  button.
button.
The  button allows panning across the zoomed plot.
button allows panning across the zoomed plot.
Note
This example is in demo/jupyter/Display_User_Interaction.ipynb and
demo/scripts/Display_User_Interaction.py.